exosome and extracellular vesicle characterization
The problem
Quantifying and characterizing exosomes and extracellular vesicles (EVs) is not an easy task. Their small size, low protein content and habit of co-purifying with lipoproteins causes a world of pain for most analytical platforms. Samples commonly contain relatively low concentrations of vesicles, making purification and concentration of the starting material essential before you can even get started on analysis.
Techniques such as nanoparticle tracking analysis (NTA) and tunable resistive pulse sensing (TRPS) fall flat in complex samples. Their non-specific approach means they pick up on all the non-vesicular particles present, affecting the specificity and sensitivity of the results. Antibody-based technologies including fluorescent NTA (fNTA) and nano-flow cytometry allow for more specific exosome characterization but have their limitations. fNTA brings marker-based specificity to NTA, but suffers badly from photobleaching, making subpopulation analysis an uphill battle. Nano-flow cytometry, like all techniques, requires sample purification and concentration, and struggles to reliably size exosomes under 60 nm.
The right tool for the job
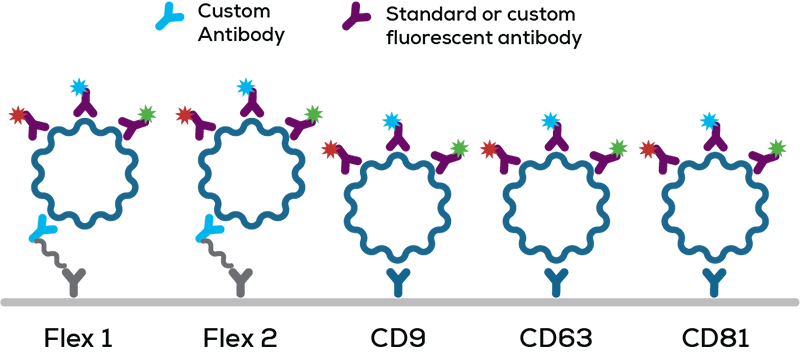
Leprechaun pulls together fluorescence imaging and single particle interferometry to provide complete characterization of exosomes from any sample type, without the need for purification. Only measure the EVs you care about with Leprechaun's Luni consumable. Using an antibody microarray the Luni uses tetraspanin antibodies to specifically snag your exosomes from solution, before fluorescent antibodies confirm you're looking at the right stuff. This antibody tag team stops contaminating particles and lipoproteins from skewing your results. Nail down exosome concentration, size and phenotype from <25 µL per sample with single vesicle sensitivity.

The proof
Exosome concentrations galore
Measure exosome concentration all the way down to 5x105 particles/mL, no need to worry about missing the rarest EVs in the most complex samples. Leprechaun combines data from three fluorescence channels to build up a picture of the proteins present on each exosome. Add in your own fluorescent antibodies and get the lowdown on the concentration of EVs that express your custom marker.

If you need to suss out how many of your exosomes have the correct cargo just fix, permeabilize and probe for your target. Leprechaun will let you know how many exosomes have the correct contents and which surface markers your cargo colocalizes with.


Size even the smallest EVs
Using single particle interferometry (SP-IRIS) enables Leprechaun to measure high resolution size distributions of exosomes as small as 35 nm, opening up a world of small EVs other platforms can't touch. Why settle for sizing based on bulk particle analysis when you can get exosome-by-exosome data with Leprechaun? Measuring each EV individually lets you combine sizing data with phenotypic info from fluorescent antibody staining, to build up a size profile for any EV subpopulation you want.

Flex your EV
Looking for a really rare exosome population? Flex Lunis mean you can create custom assays from the comfort of your own lab. Leprechaun clears the way for you to switch out the standard tetraspanin antibodies and pull-down exosomes on a marker of your choice to enrich for rare EVs.

Leprechaun
Leprechaun is the only system that hunts down titer and structure for viruses and exosomes without worrying about sample purity. Triple-check lentivirus particles to be sure they are the right size, have the right structure and contain RNA - in both crude and pure samples. Uncover exosome concentration and phenotype in everything from cell culture to biofluids. Make your own luck and follow Leprechaun straight to the viral titer or exosome concentration you've been looking for, without the noise from trickster particles throwing you off the trail.
Want more info?
Want to learn more about how Leprechaun hands over exosome concentration, size and phenotype info without breaking a sweat?