Differential Scanning Fluorimetry
Quickly characterize protein stability for construct and formulation screening
Speed-up stability studies
With differential scanning fluorimetry (DSF), you can quickly screen through many different proteins or formulation candidates to find the most stable ones. Instead of waiting days or weeks for an accelerated, high temp isothermal study, or even longer for a real-time stability study, DSF gets you stability answers in just a few hours by evaluating how protein conformation changes in response to a thermal ramp.
Make the most of fluorescence
Differential scanning fluorimetry assays work by measuring changes in fluorescence as a protein’s conformation changes. The fluorescence can be that of amino acids in the protein (the ‘intrinsic’ fluorescence of the aromatic residues, mainly tryptophan) or from an added reporter fluorophore such as SYPRO® Orange.
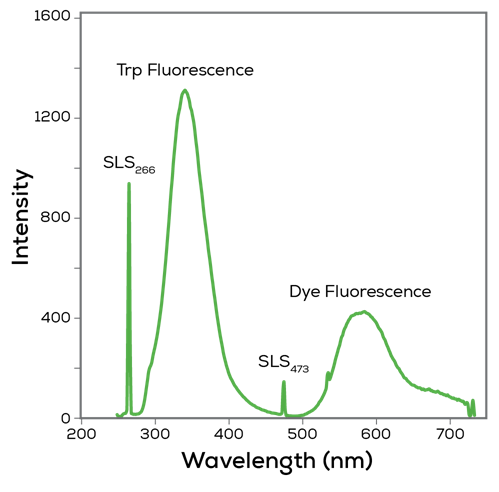
See the change
For proteins, the emitted fluorescence depends on the local environment around the fluorescent amino acids. The fluorescence spectrum of a folded protein with highly non-polar hydrophobic pockets will look different from the spectrum of an unfolded protein which has its fluorescent amino acids exposed to a more polar aqueous environment.
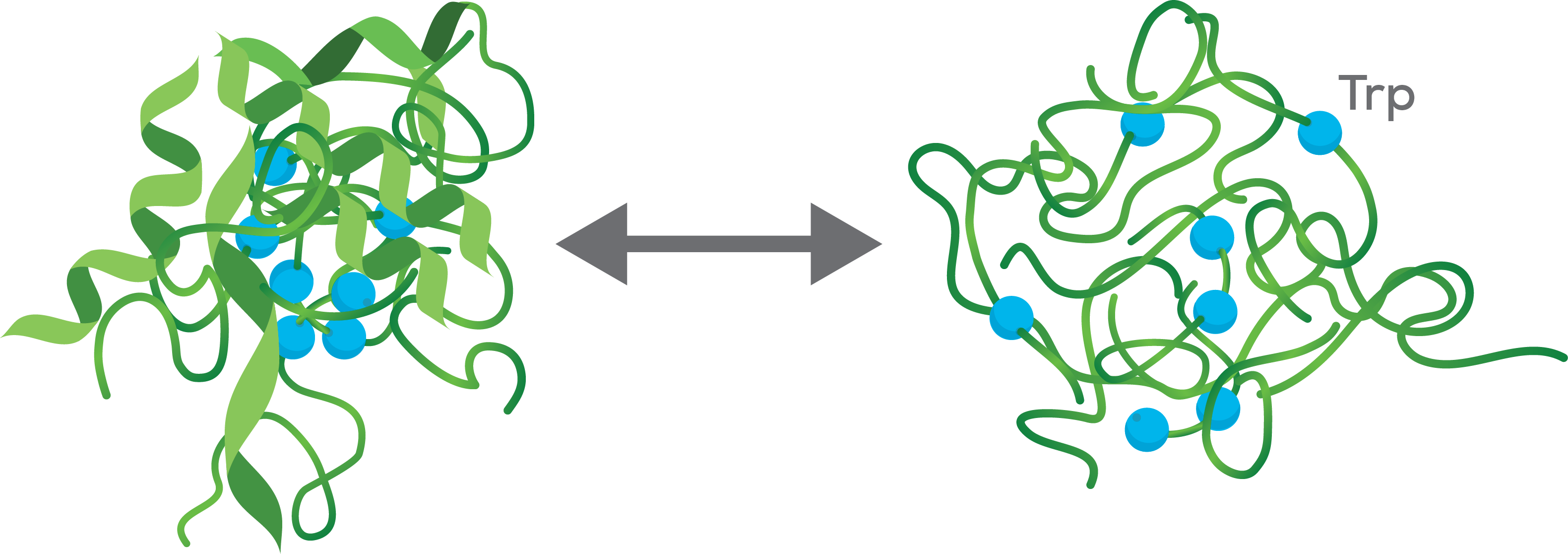
Turn up the heat
The emission spectrum of the protein will change when it melts during exposure to heat. With differential scanning fluorimetry, you are measuring changes in fluorescence as a protein unfolds in response to the increasing temperatures in a thermal ramp.
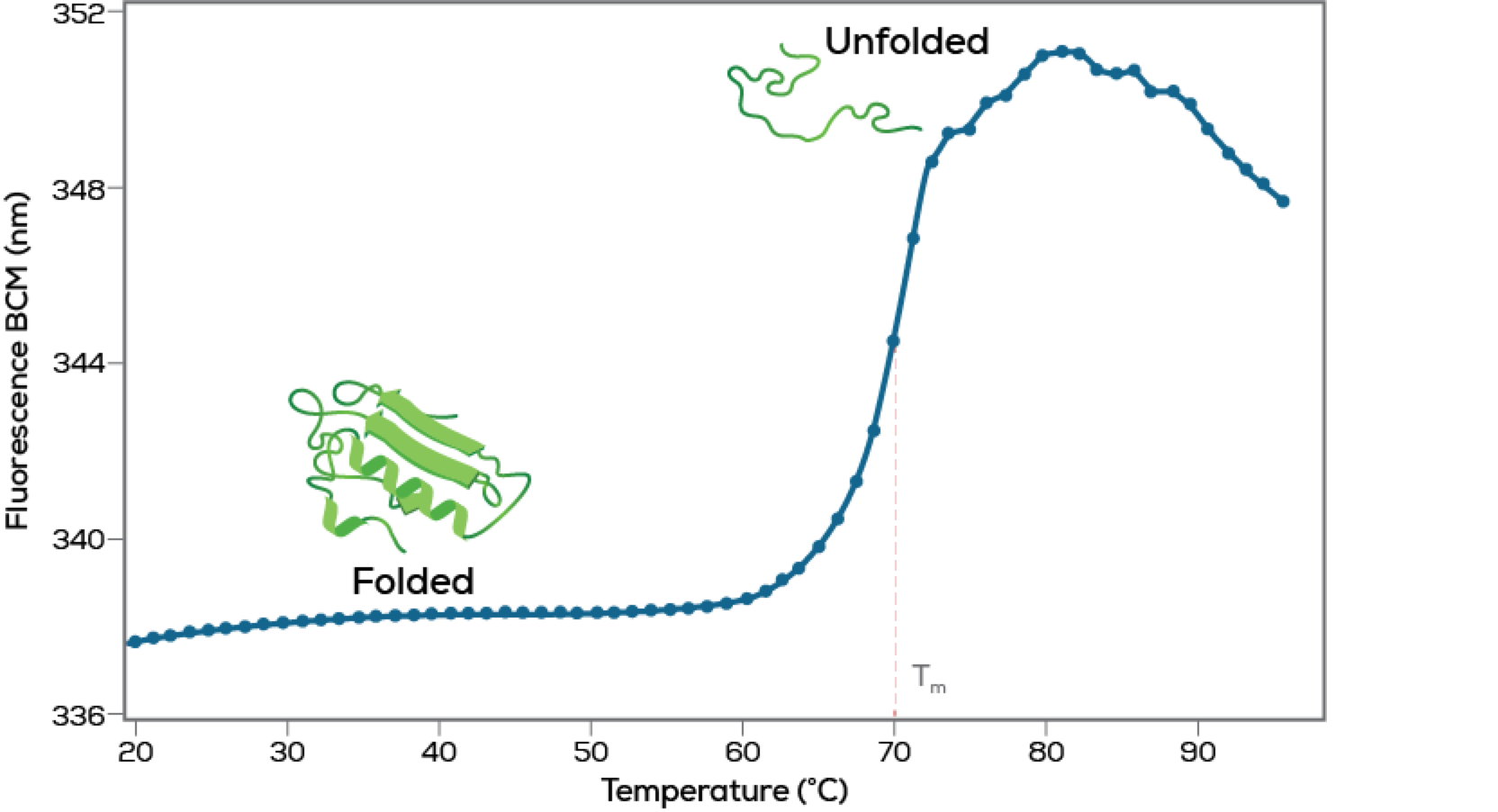
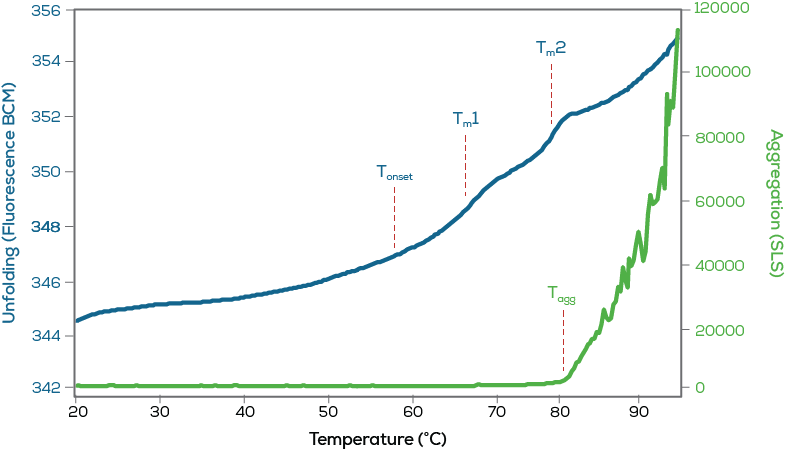
Identify inflection points
The onset temperature (Tonset) is when proteins begin to unfold. The other inflection points (Tm1, Tm2) are the temperatures at the mid-point of unfolding events.
As an added datapoint in a thermal ramp experiment, static light scattering (SLS) identifies the aggregation temperature (Tagg) of the protein. Seeing Tm and Tagg in conjunction allows identifying which unfolding events lead to aggregation.
Make the most of full-spectrum fluorescence detection
Full-spectrum fluorescence is about more than just having the best signal-to-noise: it’s a key tool to figuring out when something is wrong with your DSF assay and to having a back-up plan. Small proteins might not have fluorescent amino acids, a buffer may also show fluorescence or it might quench the intrinsic protein fluorescence signal—all things that full-spectrum detection helps identify.
Thankfully, differential scattering fluorimetry analysis is not limited to using the sample’s intrinsic signal. The technique can also utilize fluorescent dyes that track protein unfolding, which is an option only available in systems that can read their emission range. Further, when planning stability work with viral vectors, dyes sensitive to nucleic acids can be used with viral vectors like AAV to detect when genomes begin leaking out of capsids.

Aunty
Aunty combines DSF differential scattering fluorimetry, static light scattering, and dynamic light scattering measurements of samples in 96-well plates to deliver results for protein unfolding and aggregation screening, all recorded from the same sample. Add in customizable analysis across full-spectrum fluorescence detection and you’ve got a flexible tool that is ready to answer any protein stability question with unrivalled speed and data quality. Seeing the whole story on when proteins unfold and aggregate reveals exactly what’s going on with protein samples. Aunty also provides insights into long term stability or colloidal stability (B22, kD and G22), making it the queen of protein stability.
FAQs
Ready for more?
Biologics researchers can now use the right tool for protein stability studies with an instrument that combines differential scanning fluorimetry (DSF), static light scattering (SLS), and dynamic light scattering (DLS). Have a question or ready to find out more?